Yen Ting LinPostdoc
CNLS/t-6 Nonequilibrium statistical mechanics and biological physics 
Office: TA-3, Bldg 1690, Room 138
Mail Stop: B258
Phone: (505) 606-8038
Fax: (505) 665-2659 yentingl@lanl.gov
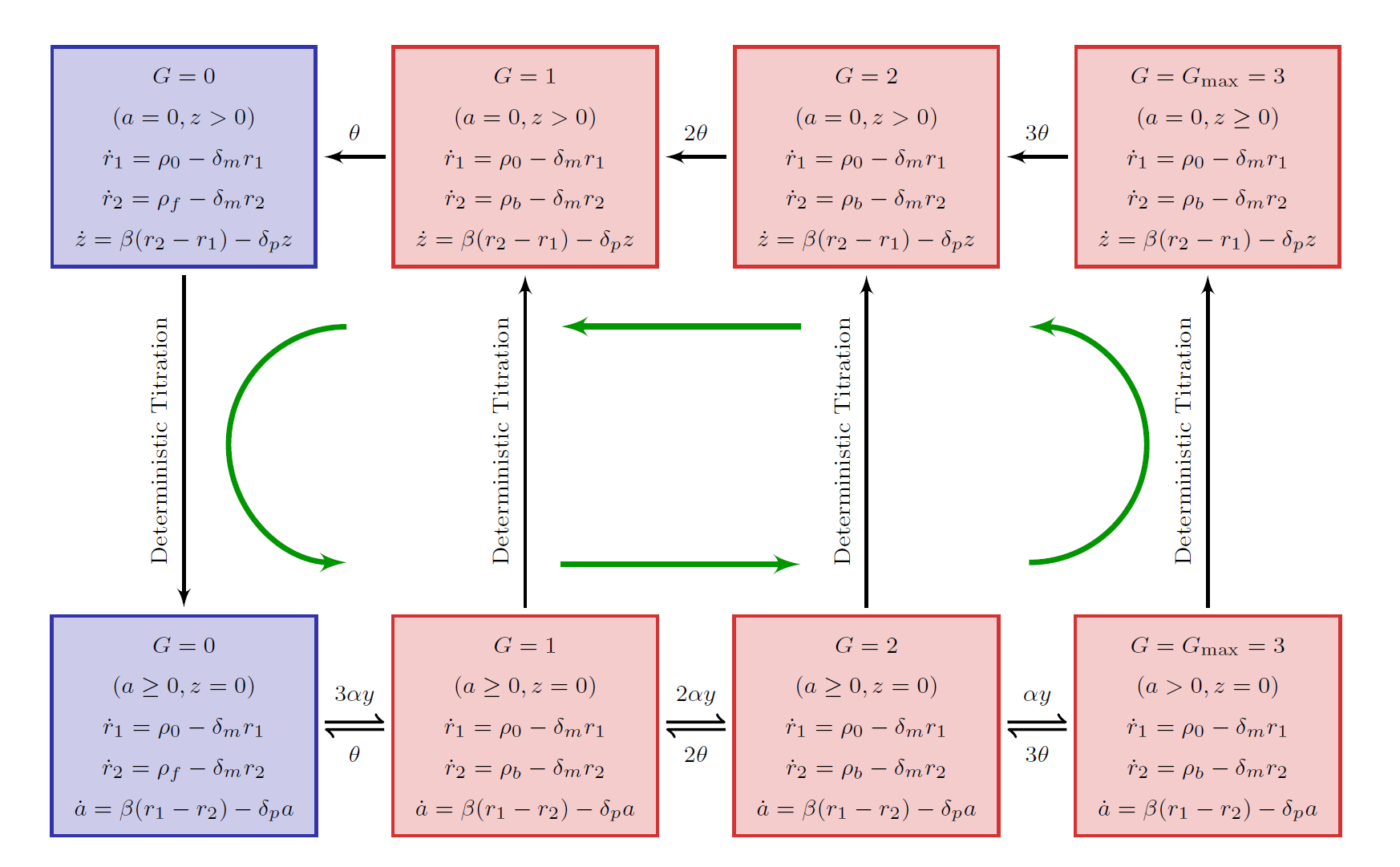
home page Research highlight- Y. T. Lin and N. E. Buchler, Efficient analysis of stochastic gene dynamics in the non-adiabatic regime using piecewise deterministic Markov processes. J. R. Soc. Interface 15: 20170804 (2018)
- Y. T. Lin, Song Feng, and W. S. Hlavacek, Scaling methods to accelerate stochastic simulation for large and realistic chemical reaction networks (in final preparation for submission)
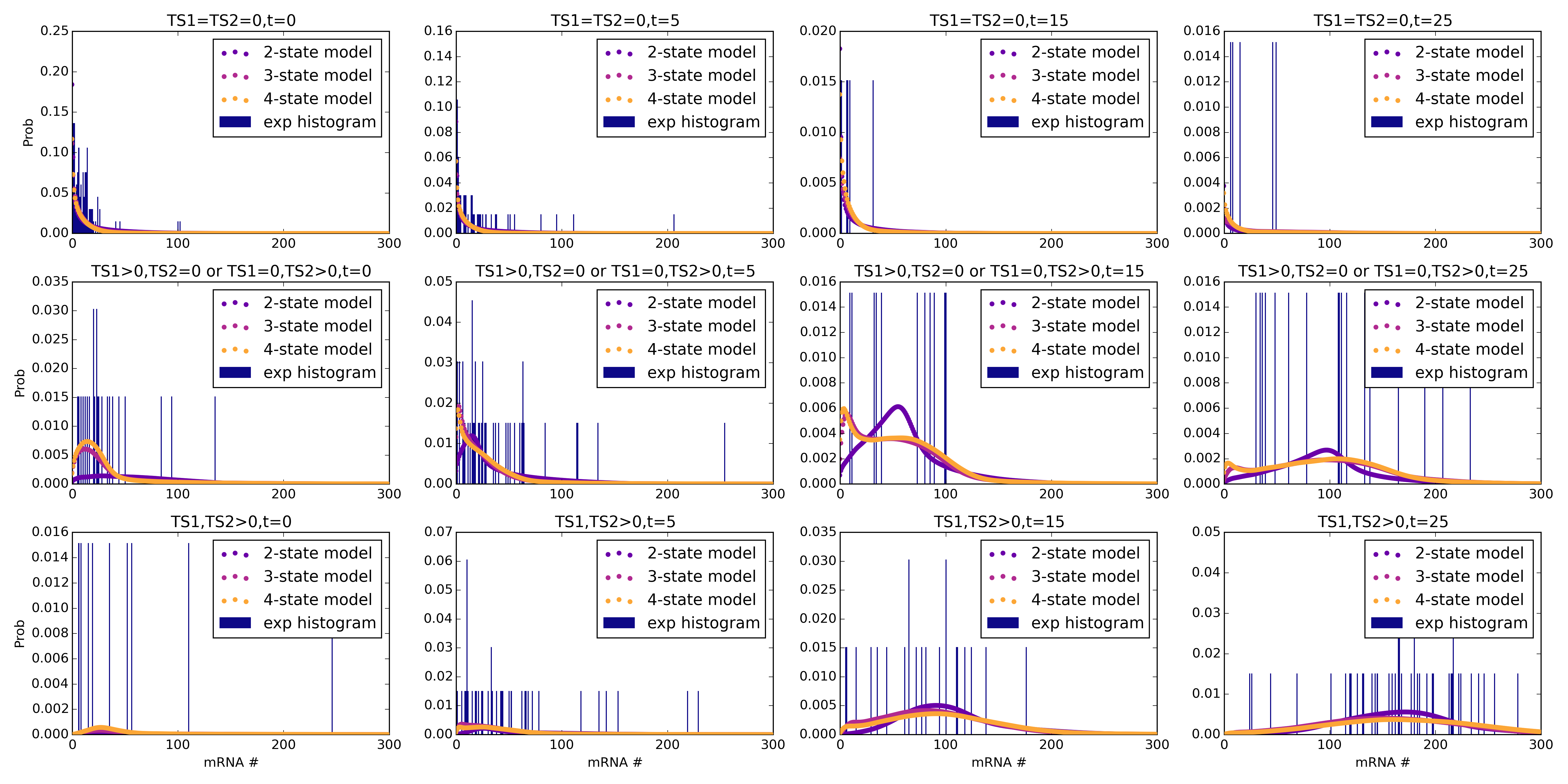
- Y. T. Lin and N. E. Buchler, Accelerated Bayesian inference of gene expression models from snapshots of single-cell transcripts (arXiv:1812.02911 [q-bio.QM])
|  | Educational Background/Employment:- B.Sc. (2004) Department of Physics, National Taiwan University, TW
- M.Sc. (2006) Theoretical physics, Department of Physics, National Taiwan University, TW
- Ph.D. (2013) Department of Physics, University of Michigan -- Ann Arbor, USA
- Employment:
- Postdoctoral Fellow (2013-2014), Department of Biological Physics, Max Planck Institute for the Physics of Complex Systems, DE
- Postdoctoral Research Associate (2015-2016), Theoretical Division, The University of Manchester, UK
Research Interests: - Non-equilibrium statistical physics
- Applied stochastic processes
- Biological physics
- Asymptotic methods
- Uncertainty quantification
- Statistical inference
- Computational system biology
Selected Recent Publications: Here is my Google scholar page and CV
- A. Shirin, I. Klickstein, S. Feng, Y. T. Lin, W. S. Hlavacek, and F. Sorrentino, Optimal Drug Cocktails and Drug Dosages to Control Autophagy, Scientific Reports, in press (2019)
- L.-F. Chen, Y. T. Lin, D. A. Gallegos, M. F. Hazlett, M. Gómez-Schiavon, M. G. Yang, B. Kalmeta, A. S. Zhou, L. Holtzman, C. A. Gersbach, J. Grandl, N. E. Buchler, and A. E. West, Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes, Cell Reports, 26, 1174-1188 (2019)
- K. E. Erickson , O. S. Rukhlenko, M. Shahinuzzaman, K. P. Slavkova, Y. T. Lin, R. Suderman, E. C. Stites, M. Anghel, R. G. Posner, D. Barua, B. N. Kholodenko, W. S. Hlavacek, Modeling cell line-specific recruitment of signaling proteins to the insulin-like growth factor 1 receptor, PLoS Computational Biology, 15(1): e1006706 (2019)
- W. Huang, Y. T. Lin, D. Frömberg, J. Shin, F. Jülicher, and V. Zaburdaev, Exactly solvable dynamics of forced polymer loops, New J. Phys. 20, 113005 (2018)
- Y. T. Lin, N. W. Lemons, L. A. Chylek, and W. S. Hlavacek, Using equation-free computation to accelerate stochastic simulation of chemical kinetics, J. Phys. Chem B, 122(24), 6351-6356 (2018)
- P. Bokes, Y. T. Lin, A. Singh, High Cooperativity In Negative Feedback Can Amplify Noisy Gene Expression, Bull. Math. Biol. (2018)
- Ryan Suderman, Eshan D. Mitra, Yen Ting Lin, Keesha E. Erickson, Song Feng1,2, and William S. Hlavacek, Generalizing Gillespie’s Direct Method to Enable Network-Free Simulations, Bull. Math. Biol. online first (2018)
- Y. T. Lin, P. Hufton, E. Lee, D. Potoyan, A stochastic and dynamical view of pluripotency in mouse embryonic stem cells, PLoS Computational Biology 14(2): e1006000 (2018)
- Y. T. Lin and N. E. Buchler, Efficient analysis of stochastic gene dynamics in the non-adiabatic regime using piecewise deterministic Markov processes. J. R. Soc. Interface 15: 20170804 (2018)
- P. Hufton, Y. T. Lin and T. Galla, Phenotypic switching of populations of cells in a stochastic environment, J. Stat. Mech. 023501 (2018)
- Y. T. Lin, E. T. Y, Chang, J. Eatock, T. Galla, R. H. Clayton, Mechanisms of stochastic onset and termination of atrial fibrillation studied with a cellular automaton model, J. R. Soc. Interface 14: 20160968 (2017)
- P. Hufton, Y. T. Lin, T. Galla, and A. J. McKane, Intrinsic noise of population dynamics in randomly switching environments, Phys. Rev. E 93 052119 (2016)
- E. T. Y, Chang, Y. T. Lin, T. Galla, R. H. Clayton, and J. Eatock, A Stochastic Individual-Based Model of the Progression of Atrial Fibrillation in Individuals and Populations, PLoS ONE 11(4): e0152349 (2016)
- Y. T. Lin and C. R. Doering, Gene expression dynamics with stochastic bursts: construction and exact results for a coarse-grained model, Phys. Rev. E 93, 022409 (2016)
- Y. T. Lin and T. Galla, Bursting noise in gene expression dynamics: Linking microscopic and mesoscopic models, J. R. Soc. Interface 13: 20150772 (2016)
- Y. T. Lin, D. Frömberg, W. Huang, P. Delivani, M. Chacón, I. M. Tolic, F. Jülicher, and V. Zaburdaev, Pulled polymer loops as a model for the alignment of meiotic chromosomes, Phys. Rev. Lett. 115 208102 (2015)
- J. Eatock, Y. T. Lin, E. T. C. Chang, T. Galla, R. H. Clayton, Assessing Measures of Atrial Fibrillation Clustering via Stochastic Models of Episode Recurrence, Proceedings of Computing in Cardiology (2015)
- C. Weber, Y. T. Lin, N. Biais, and V. Zaburdaev, Formation and dissolution of bacterial colonies, Phys. Rev. E 92 032704 (2015)
- J. Taktikos, Y. T. Lin, H. Stark, N. Biais, and V. Zaburdaev, Pili-induced clustering of N. gonorrhoeae bacteria, PLoS ONE 10(9): e0137661 (2015)
- Y. T. Lin, H. Kim, and C. R. Doering, Demographic stochasticity and the evolution of dispersion II. Spatially inhomogeneous environments, J. Math. Biol. 70(3) 679-707 (2015, accepted in 2014, submitted in 2013).
- Y. T. Lin, H. Kim, and C. R. Doering, Demographic stochasticity and the evolution of dispersion I. Spatially homogeneous environments,
J. Math. Biol. 70(3) 679-707 (2015, accepted in 2014, submitted in 2013).
- Y. T. Lin, H. Kim, and C. R. Doering, Features of fast living: On the weak selection for longevity in degenerate birth-death processes, J Stat. Phys. 148(4) 646-662 (2012)
- E. Khain, Y. T. Lin, and L. M. Sander, Fluctuations and stability in front propagation, Euro. Phys. Lett., 93 28001 (2011)
|



